DNA Search Engine for Learning, From Lists to Graphs
Input
Modified
A DNA search engine replaces lists with graph-native matching MetaGraph proves scalable, fast, accurate, and low-cost sequence search Education should adopt path-based search with strong data governance

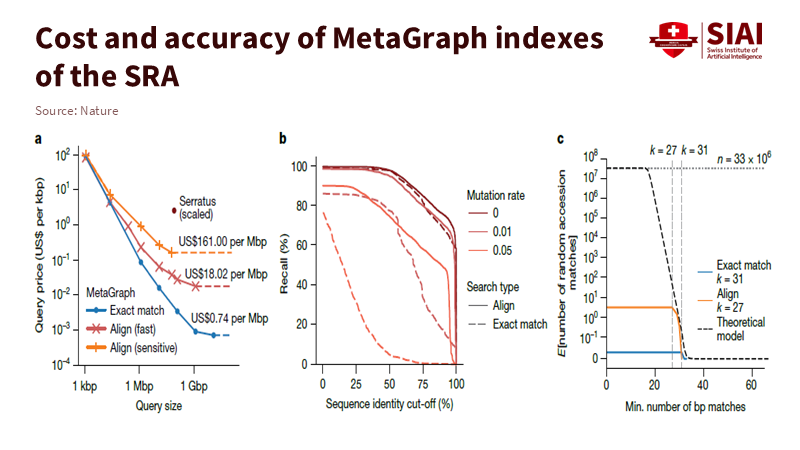
Public archives now hold vast amounts of genetic data measured at the petabase scale. A new DNA search engine, MetaGraph, can navigate this data and provide precise results quickly. In the Nature study announcing it, MetaGraph makes 18.8 million unique DNA and RNA sequence sets from seven primary sources searchable in full text. This turns previously isolated reads into one coherent index. Instead of ranking pages, it matches many slight overlaps simultaneously and traces them across a network. This change, from focusing on single-point hits to multi-point paths, could reshape how we approach learning search. Biology faced a challenge with the size and noise of data, which it addressed by changing the search structure. Education is experiencing a similar issue: repeated content, messy tags, and weak signals that only make sense when combined. Just as genomes need a graph for effective searching, our educational materials do too. The lesson is clear: altering the structure of search changes what students can find.
Why does a DNA search engine reset the learning search?
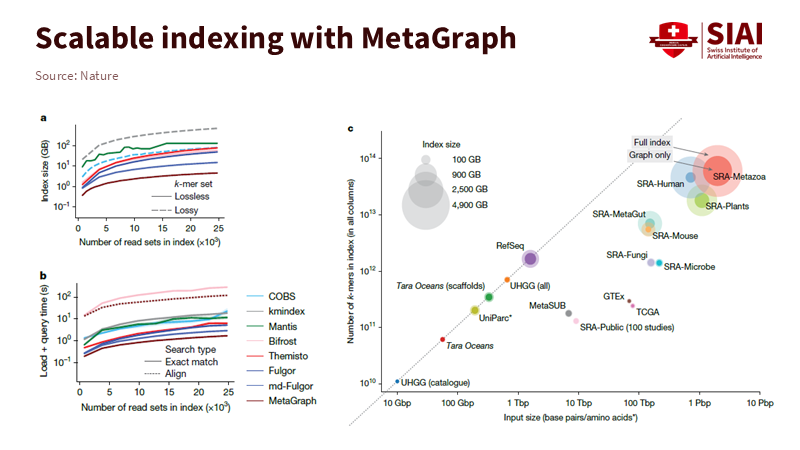
Education still operates with search methods from a decade ago. Users type a query and get a list. We scroll down, hoping the correct item appears near the top. A DNA search engine functions differently. It sees each sequence as thousands of short tokens and searches for paths that confirm each other. Many weak signals come together to form one strong answer. This method is better at handling noise and capturing structure. The order of sequences matters, as does the order in scholarship. Methods lead to datasets, and datasets lead to code. A list obscures those connections, while a graph uncovers them. This is crucial because of scale. Biological repositories are not just large; they are interconnected. GenBank alone shows continuous growth across accessions and base pairs. MetaGraph’s index encompasses diverse clades, from viruses to humans, and can return results in seconds to minutes rather than hours. This speed is achievable because the engine searches overlaps instead of files. Education’s repositories may be smaller, but the underlying principles apply.
The practical benefit is that a list emphasizes one signal, such as page rank or clicks. A graph gathers many weak signals and combines them. That’s how the DNA search engine detects faint patterns across thousands of samples. A next-generation learning engine should follow this same logic. As sequence sets increase, load and query times remain manageable because queries navigate the graph rather than comparing each sequence to every other sequence. For education, we should replace “reads” with “learning objects.” We should swap “alignment” for “near-match across method and dataset.” We can search the entire campus corpus if we use overlaps and search paths instead of pages.
How a DNA search engine works (without the math)
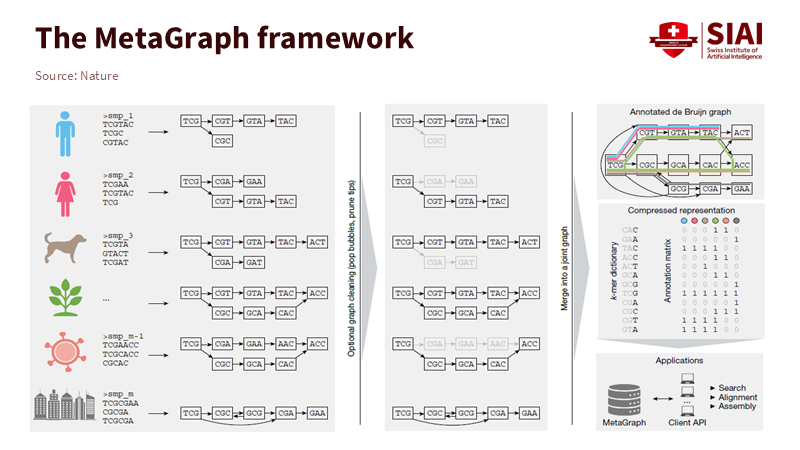
The fundamental concept comes from graph theory: the de Bruijn graph. The engine divides each sequence into overlapping k-mers. Each k-mer acts as a node. Edges connect nodes that overlap by k-1 characters. This way, the index encodes order and reuse within a single structure. A token that appears in many datasets is stored just once. Modern systems “color” nodes to indicate where each token appears and compress these colors by taking advantage of similarities along paths. The math is complicated, but the idea is straightforward: reuse overlaps to compress and then search paths to retrieve. That is how petabases become manageable indexes without losing context across projects.
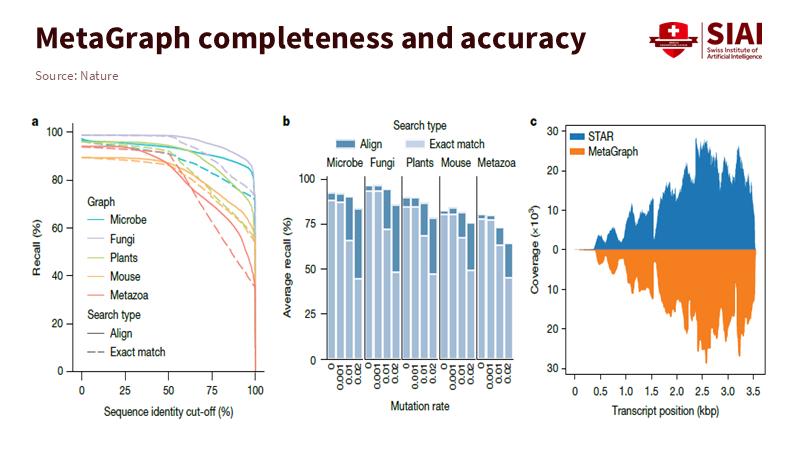
Speed means little without accuracy. MetaGraph keeps high recall even when identity cut-offs decrease and mutation rates increase. These conditions reflect real-world noise. Coverage of human transcripts shows that labels and positions are recovered without needing to realign every read against every reference. Learning search requires the same tolerance since students use imperfect words, and metadata can be disorganized. A graph can still find the correct route because it evaluates adjacent cues together, not just an exact phrase. In essence, it treats knowledge like a network instead of a shelf.
From lab discovery to classroom discovery
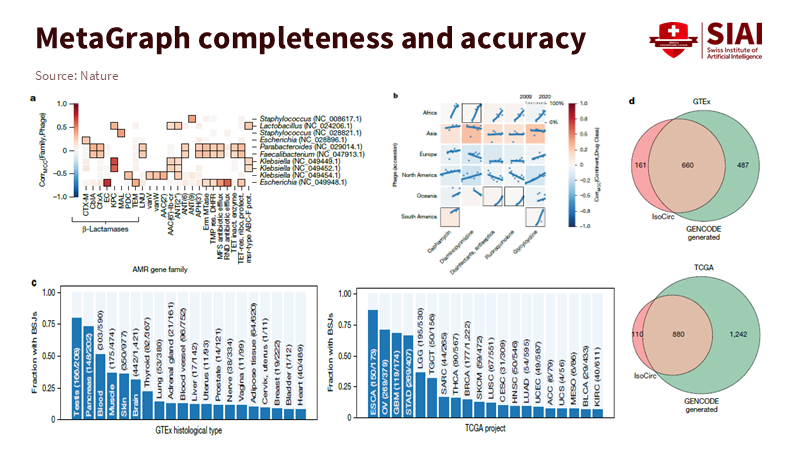
A DNA search engine does more than retrieve results. It uncovers relationships. In biology, this could mean linking antimicrobial resistance genes to phage hosts, tracking signals across cohorts, or showing overlaps between reference catalogs and clinical projects. These examples teach us a larger lesson for education. When the corpus is indexed as connected tokens, the engine provides paths that explain why items belong together. A student searching for “panel data with fixed effects” should not be met with a list of links. They should see a route that offers a brief explanation, a runnable notebook, a small dataset, and a task, with each step accessible in one click. The graph already contains these links; we need to highlight them.
This change will transform how we teach and manage reuse. First, discovery by method. Index methods, datasets, notebooks, and assignments as tokens. Annotate their locations (course, module, license) and allow queries to navigate method-first routes. Second, reuse with clear rules. The FAIR principles—Findable, Accessible, Interoperable, Reusable—become actionable at query time when every token has a source, a license, and a persistent ID. Third, create evidence loops. When a path leads to better outcomes—such as higher scores and quicker mastery—the system strengthens that route for similar students. This technology is no longer experimental. Nature and institutional briefings outline practical performance and quick queries over public repositories. Our task is to apply the same model to learning data that, while smaller, are still interconnected.
Costs, governance, and the next five years
The financial aspect is often the first challenge. In biology, however, indexing methods that reuse overlaps incur lower costs as data expands. Even with growing repositories, the cost and latency per query remain reasonable because the system follows shared paths instead of recalculating matches from scratch. Meanwhile, the broader ecosystem indicates that sequencing and storage costs have significantly dropped. NHGRI reports a long-term decline in sequencing costs, with recent data showing full-genome costs dropping below $1,000, and even below $500, depending on the platform and sequencing depth. The takeaway for universities is straightforward: a discipline-level graph index can function on standard hardware or limited cloud budgets if the data is well annotated. The real challenge is not funding; it is design.
Governance needs to evolve with the technology. Provenance and privacy should come first. Every token and path must carry a license, source, and access rules. Students should understand why an item appears and what they can do with it. Reproducibility follows. A graph index must update as new items are added. Any graded or published work should record a time-stamped snapshot of the index to allow replay of results. Lastly, we need to focus on skills. Faculty do not need to become graph experts. Still, we do need to promote graph literacy in teacher training and library programs—covering nodes, edges, paths, colors, and the importance of reusing materials. Short modules can teach the essentials and connect them to the campus index. This approach is feasible and sets the stage for applications in other fields where DNA-style graph indexing is relevant, such as medical imaging, materials synthesis, environmental sensor sequences, and even legal clauses arranged as overlapping tokens. The same pattern applies: index overlaps, search paths, return routes.
We began with a clear assertion. A DNA search engine makes petabases searchable by changing the search structure. It replaces lists with graphs and single-point matches with multi-point paths. This transformation benefits not just biology but also the future of education. We should create indexes that reuse overlaps. Let queries explore relationships and deliver paths—not piles—while showing the rationale for each step. We must support this with strong guidelines on provenance, privacy, and reproducibility. In five years, students should type a question and see the next step in their path, with reasoning attached. Biology demonstrates that scale and noise do not hinder discovery; they require structure. By integrating that structure into our services now, we can find an ally in learning rather than a barrier. The model is available, the costs are decreasing, and the approach has been validated in a more complex domain. What remains is our commitment to apply that success to everyday educational practices.
The views expressed in this article are those of the author(s) and do not necessarily reflect the official position of the Swiss Institute of Artificial Intelligence (SIAI) or its affiliates.
References
Compeau, P. E. C., Pevzner, P. A., & Tesler, G. (2011). How to apply de Bruijn graphs to genome assembly. Nature Biotechnology, 29(11), 987–991.
ETH Zurich. (2025, October). A DNA search engine.
Karasikov, M., et al. (2025). Efficient and accurate search in petabase-scale sequence repositories. Nature.
NHGRI. (2023). DNA Sequencing Costs: Data. National Human Genome Research Institute.
Our World in Data. (2024). Cost of sequencing a full human genome (curation of NHGRI data).
Sayers, E. W., et al. (2025). GenBank 2025 update. Nucleic Acids Research, 53(D1), D56–D63.
SIB Swiss Institute of Bioinformatics. (2025, October 13). A fast, accurate “sequence search engine”. News release.





















